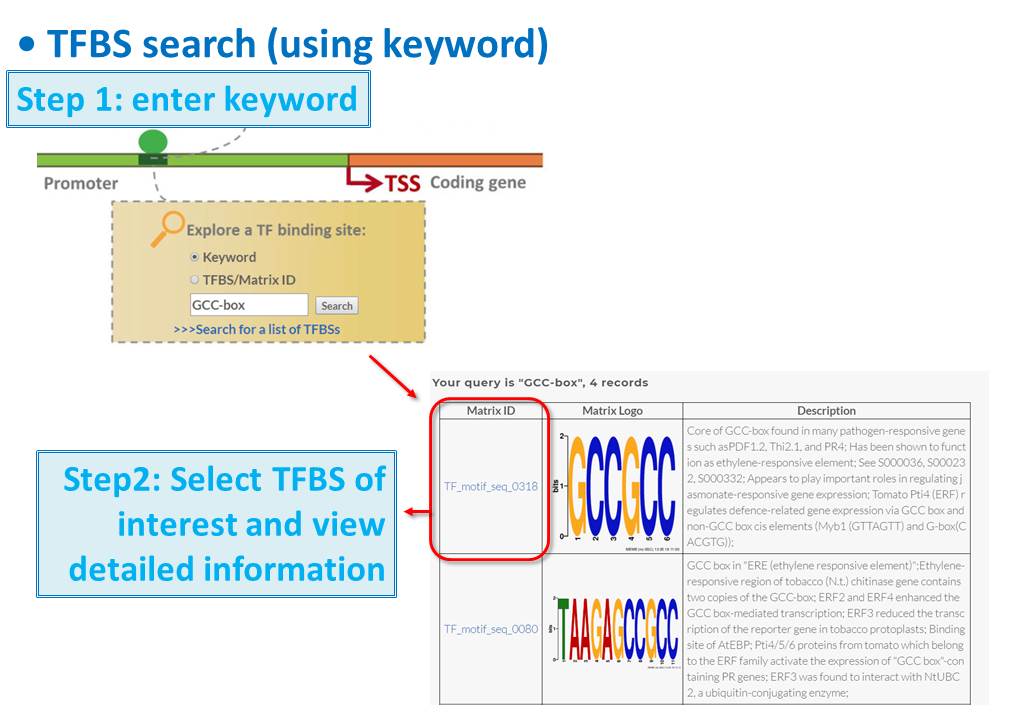
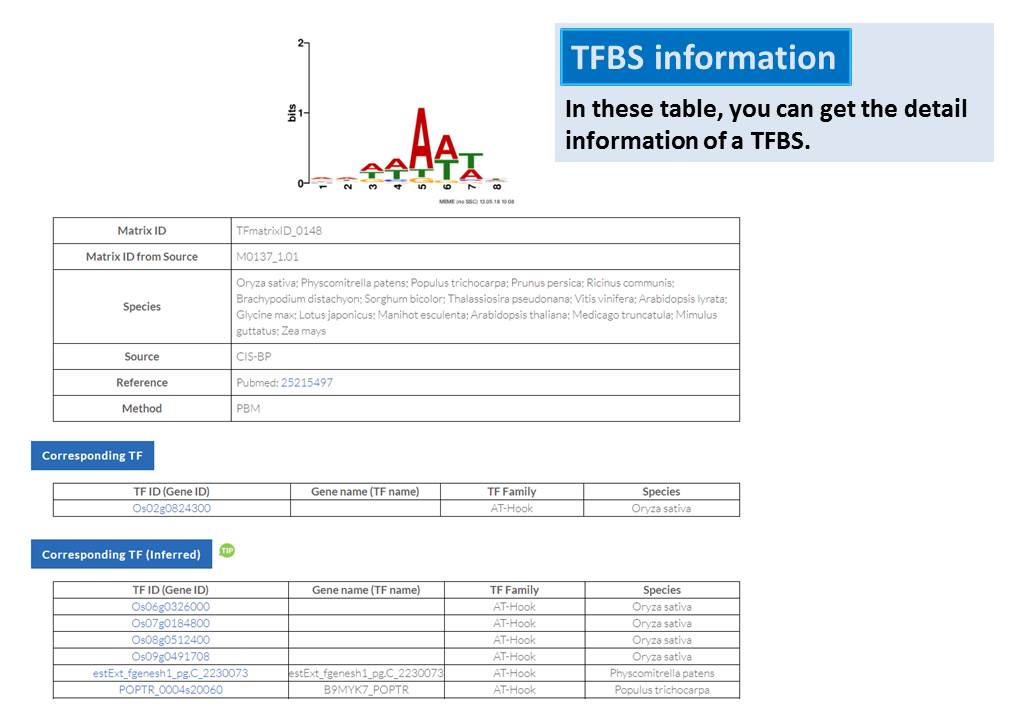
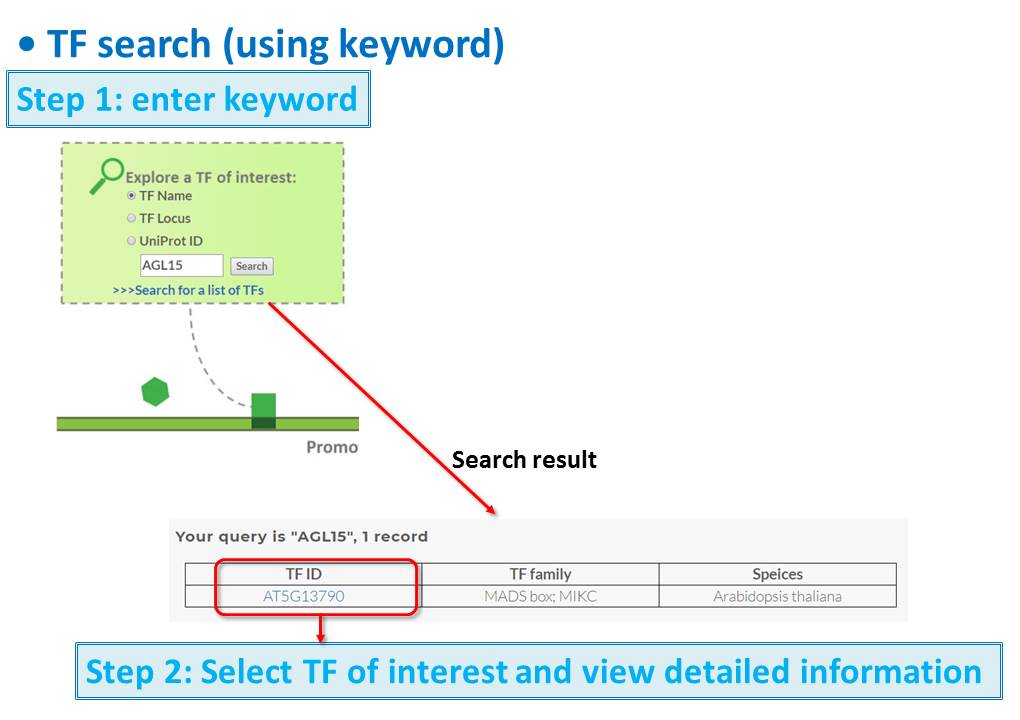
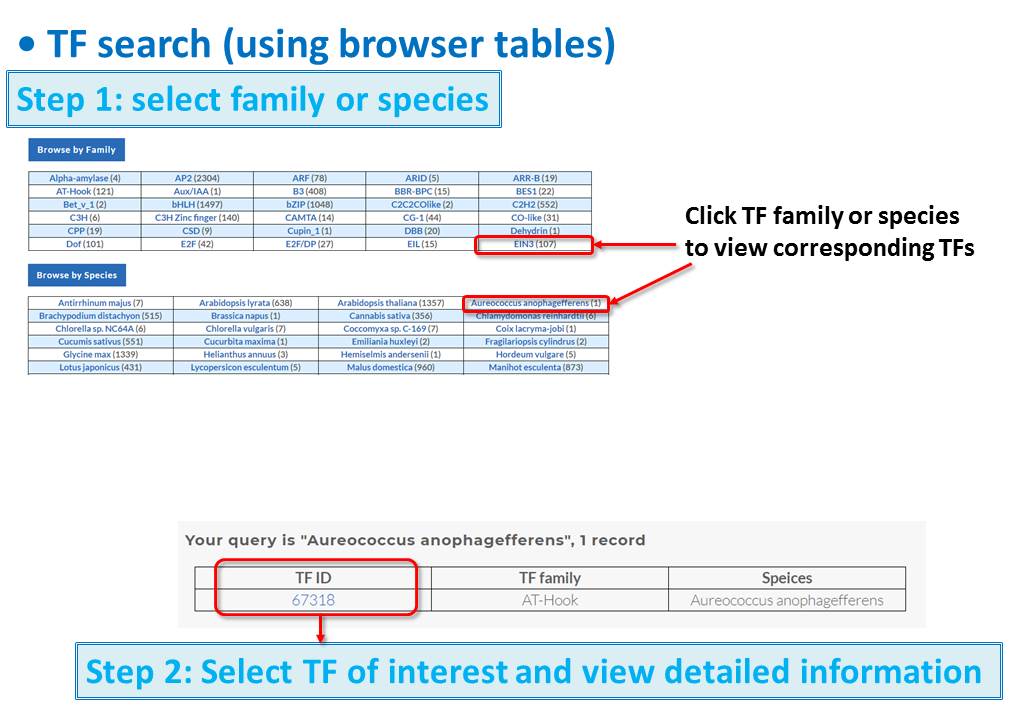
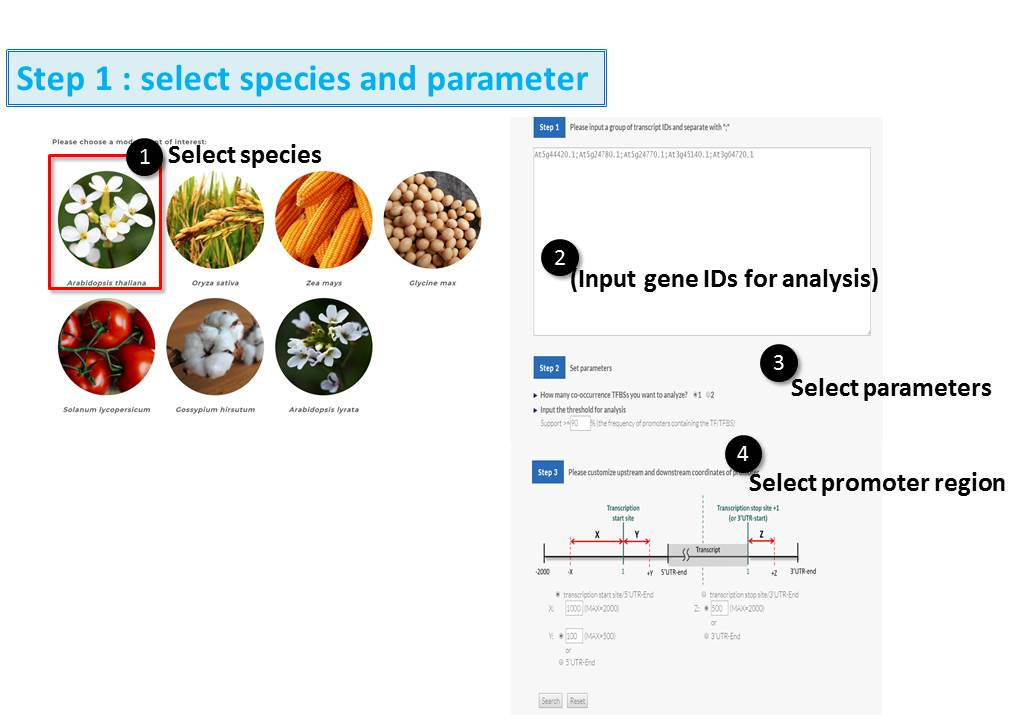
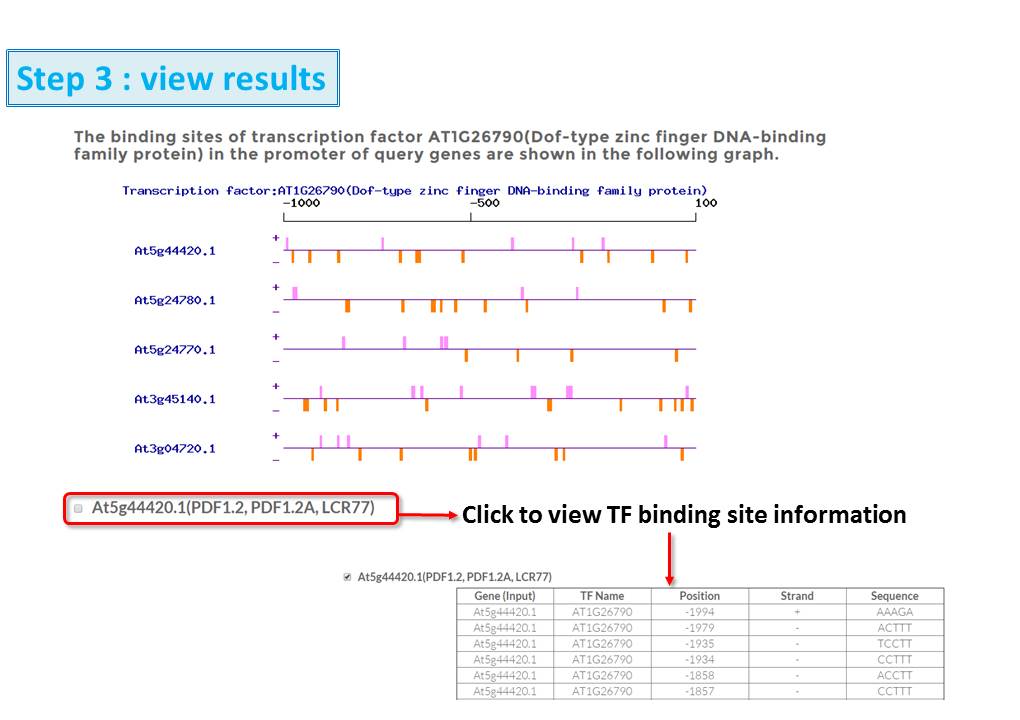
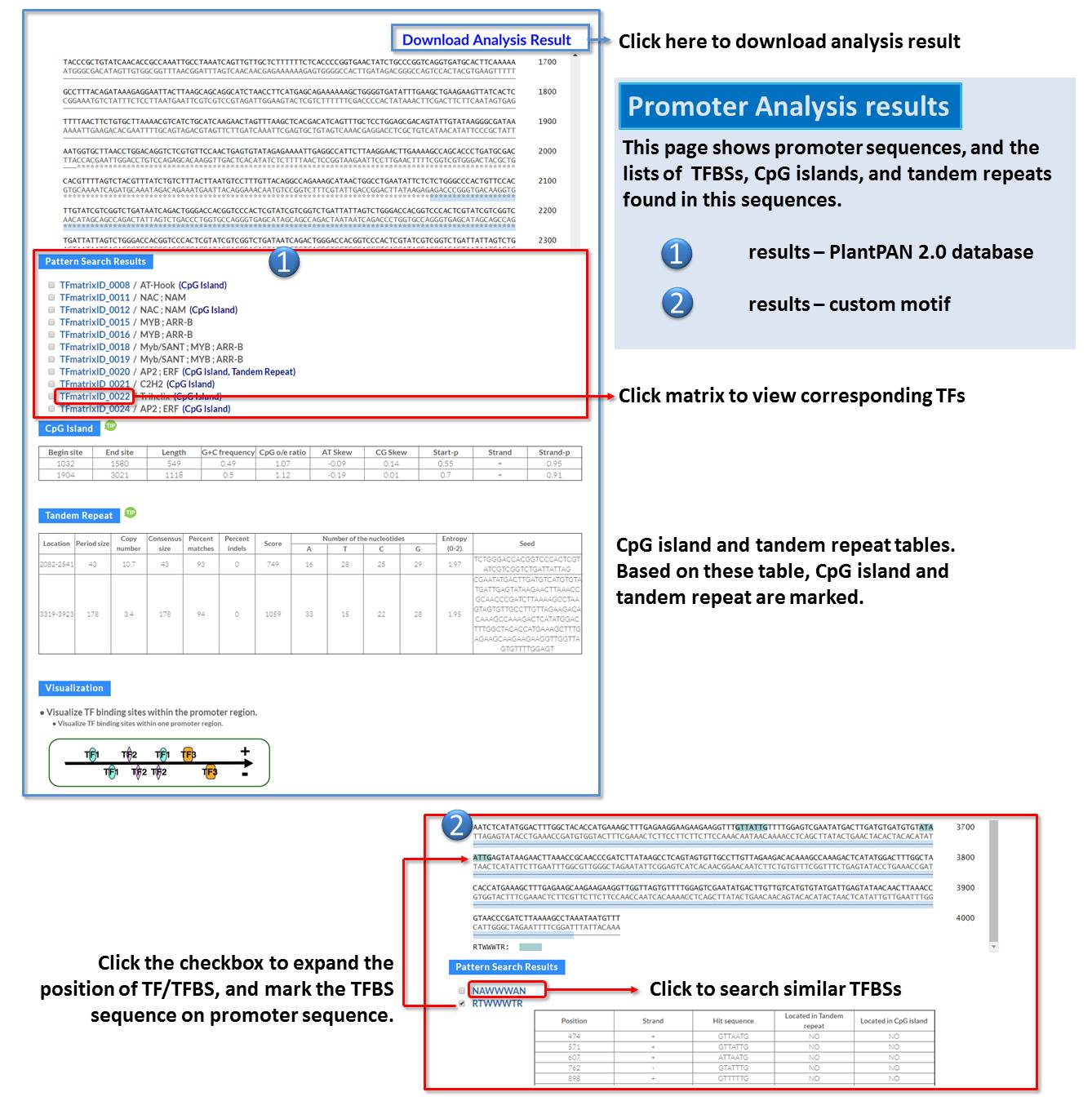
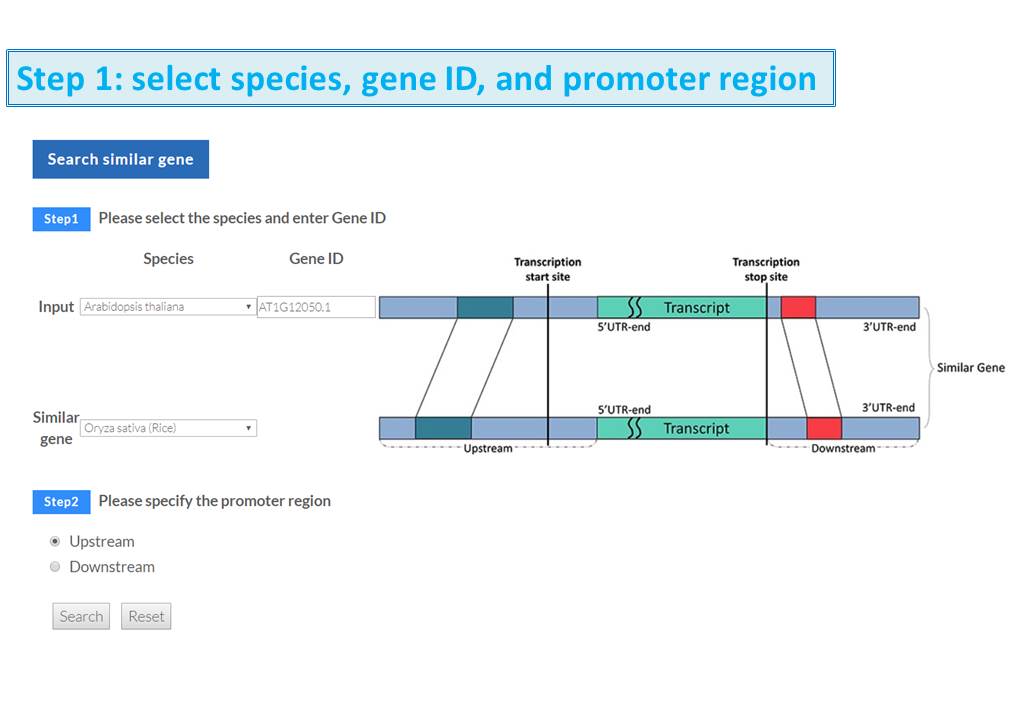
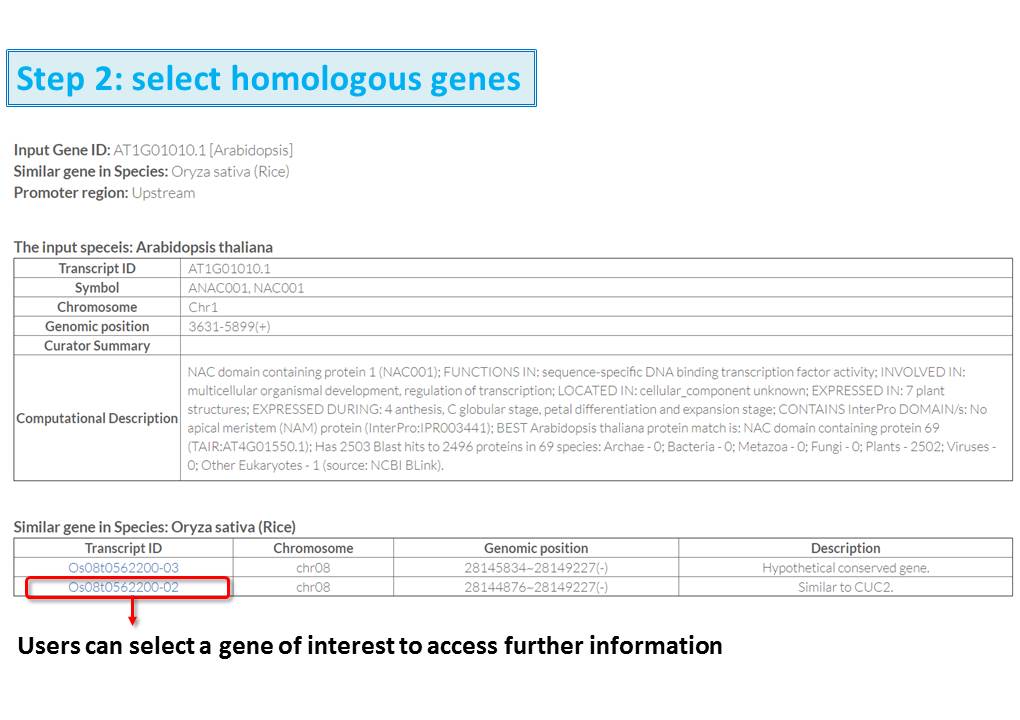
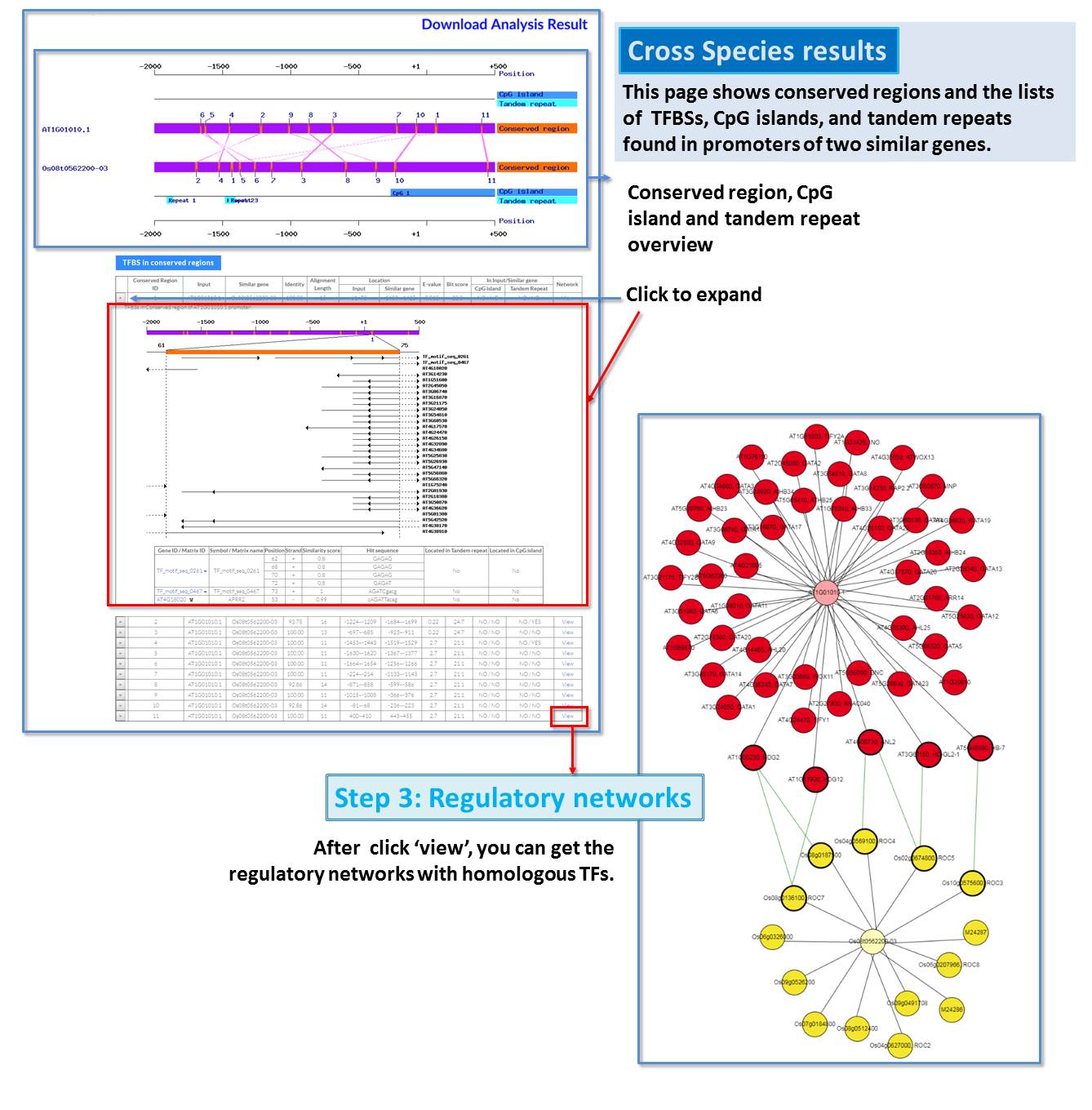
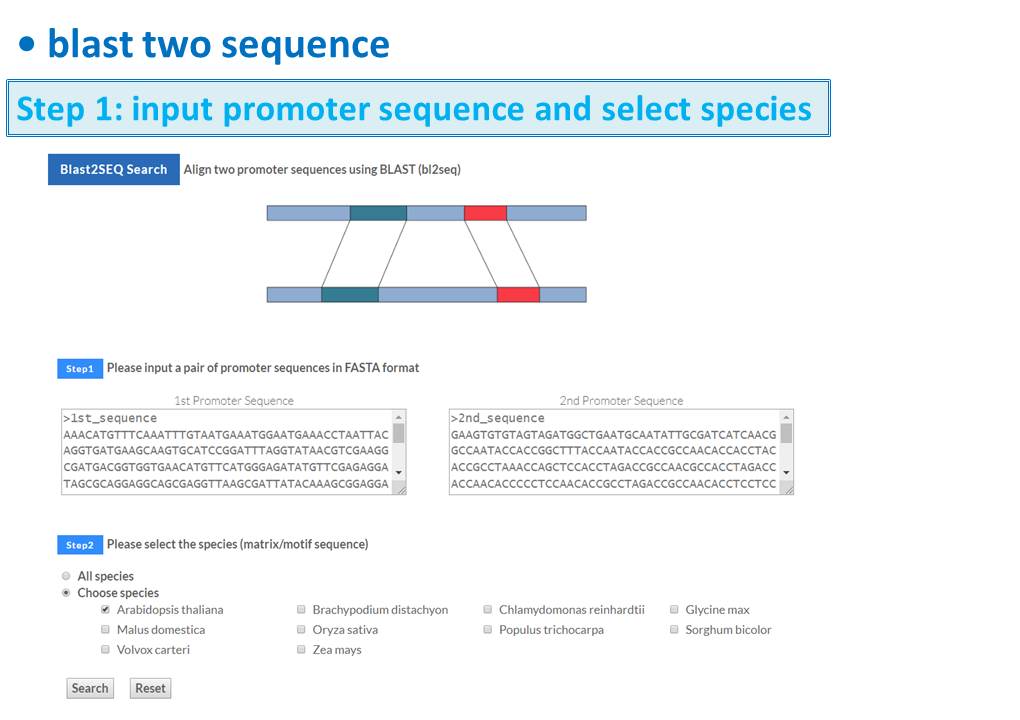
Guide - Gene Search
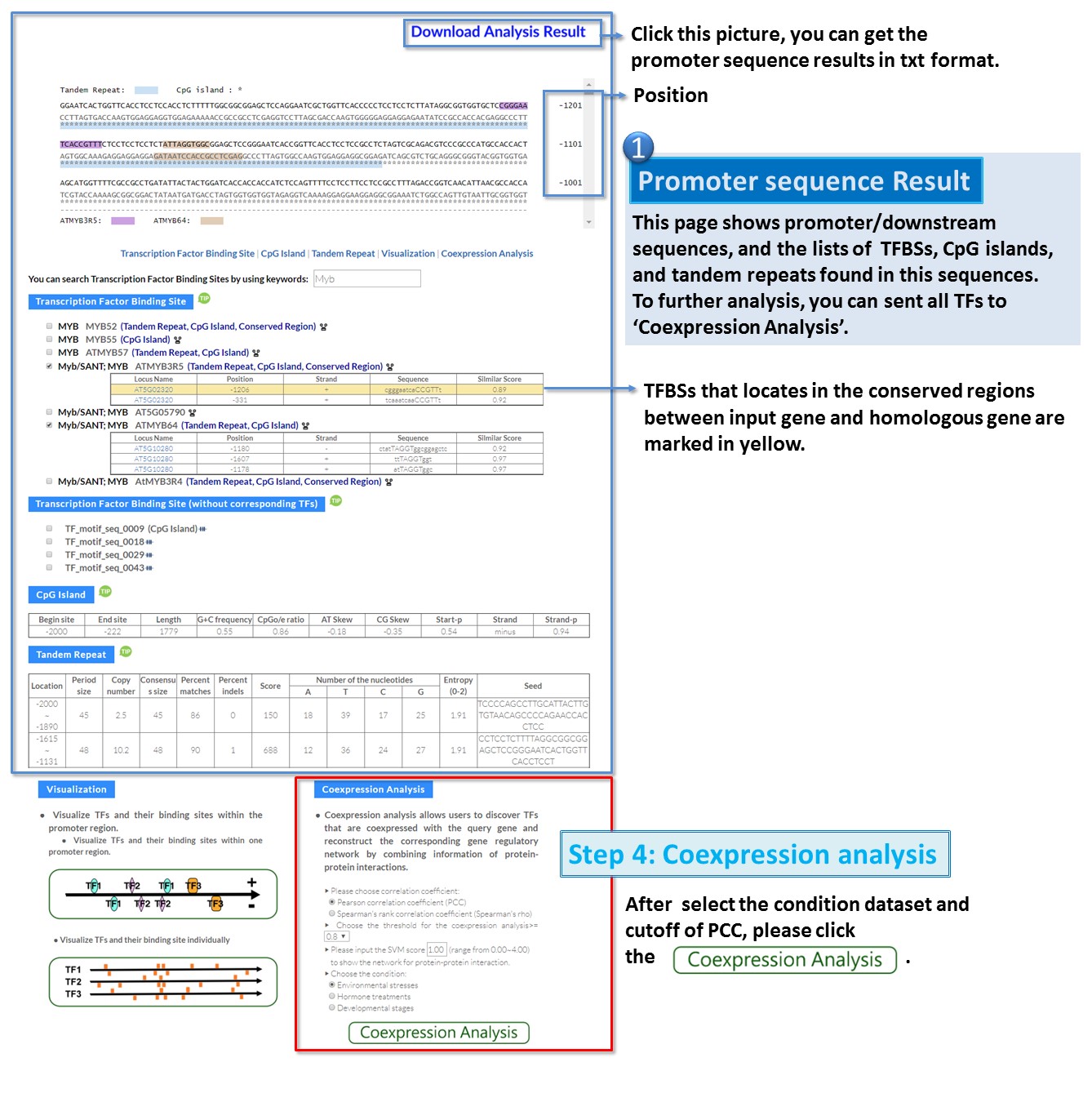
1. Identification of cis- and trans-elements of input gene.
2. Construction of gene regulatory networks by using coexpression analysis.
3. Visualization of experimental regulatory factor binding sites in JBrowse.

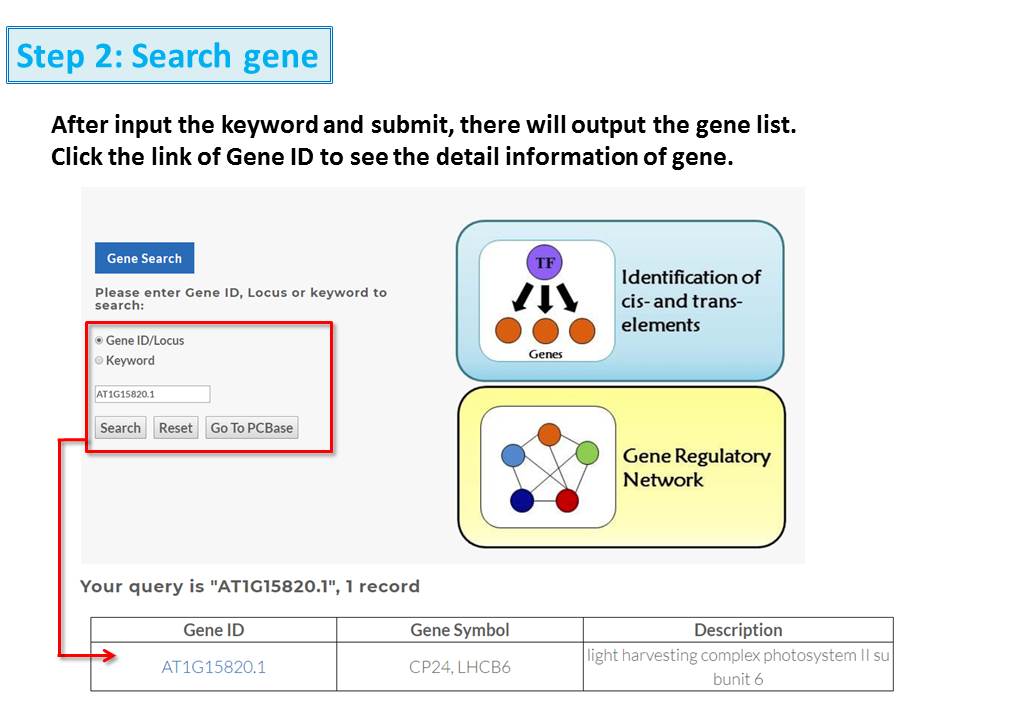


2. Construction of gene regulatory networks by using coexpression analysis.
3. Visualization of experimental regulatory factor binding sites in JBrowse.